XArray: the power of pandas for multidimensional arrays¶
Robin Wilson
@sciremotesense robin@rtwilson.com
Satellite air quality data (from the MODIS MAIAC product)¶
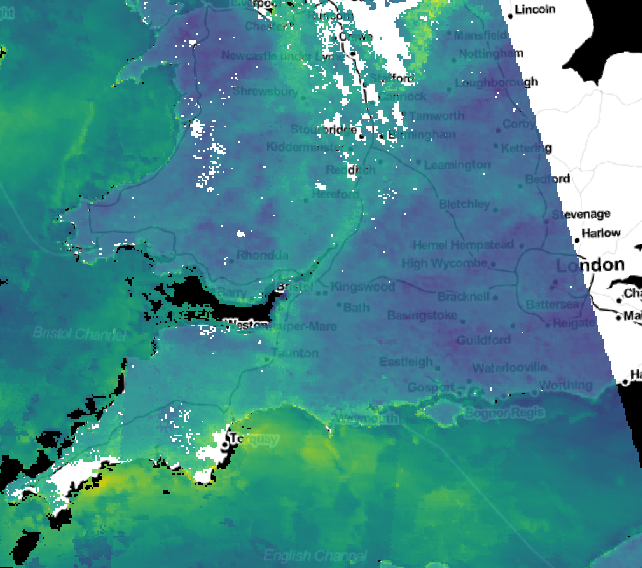
Problem¶
We have many decades of daily raster data, and want to get:
- Seasonal means and standard deviations
- Long time-series across specific points
- Individual images at specific times
The 'simple' solution¶
- Load in the image data (using
GDALorrasterio)
- Store in a large 3D
numpyarray (dimensions: X, Y and time)
- Index, slice and dice the array...
Not as easy it sounds...need to keep track of everything!
Enter XArray¶
The power of pandas for multidimensional arrays
import xarray as xr
Quick example¶
PM25 = xr.open_dataarray('/Users/robin/code/MAIACProcessing/All2014.nc')
PM25.shape
(181, 1162, 1240)
PM25.dims
('time', 'y', 'x')
seasonal = PM25.groupby('time.season').mean(dim='time')
seasonal.plot.imshow(col='season', robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x30d4aceb8>
time_series = PM25.isel(x=1000, y=1100).to_pandas().dropna()
time_series
time 2014-01-07 62.0 2014-01-17 121.0 2014-01-25 127.0 2014-02-10 135.0 2014-02-11 186.0 2014-02-18 237.0 2014-02-25 352.0 2014-02-26 293.0 2014-02-27 260.0 2014-03-01 165.0 2014-03-08 221.0 2014-03-09 221.0 2014-03-10 291.0 2014-03-13 552.0 2014-03-16 156.0 2014-03-21 159.0 2014-03-22 160.0 2014-03-23 163.0 2014-03-25 261.0 2014-03-30 362.0 2014-04-04 19.0 2014-04-09 19.0 2014-04-10 19.0 2014-04-11 19.0 2014-04-13 33.0 2014-04-14 19.0 2014-04-16 159.0 2014-04-18 53.0 2014-04-19 118.0 2014-04-24 194.0 2014-04-25 250.0 2014-05-10 154.0 2014-05-13 181.0 2014-05-14 19.0 2014-05-15 117.0 2014-05-17 239.0 2014-05-22 188.0 2014-06-02 189.0 2014-06-05 71.0 2014-06-07 107.0 2014-06-13 115.0 2014-06-14 324.0 2014-06-15 64.0 2014-06-18 132.0 2014-06-20 101.0 2014-06-21 115.0 2014-06-23 200.0 2014-06-27 162.0 dtype: float32
one_day = PM25.sel(time='2014-02-15')
one_day.plot(robust=True)
<matplotlib.collections.QuadMesh at 0x31dcdeba8>
Summary¶
seasonal = PM25.groupby('time.season').mean(dim='time')
time_series = PM25.isel(x=1000, y=1100).to_pandas().dropna()
one_day = PM25.sel(time='2014-02-20')
Introduction to XArray¶
xarray.DataArray is a fancy, labelled version of a numpy.ndarray
xarray.Dataset is a collection of multiple DataArrays which share dimensions
(A Dataset is a representation of a NetCDF file)
arr = np.random.rand(3, 4, 2)
xr.DataArray(arr)
<xarray.DataArray (dim_0: 3, dim_1: 4, dim_2: 2)>
array([[[0.006194, 0.380531],
[0.561032, 0.360739],
[0.515117, 0.300988],
[0.650297, 0.560798]],
[[0.703038, 0.762046],
[0.703794, 0.467468],
[0.19292 , 0.395919],
[0.344915, 0.874793]],
[[0.570962, 0.881131],
[0.758426, 0.582632],
[0.419739, 0.634596],
[0.720827, 0.12582 ]]])
Dimensions without coordinates: dim_0, dim_1, dim_2
xr.DataArray(arr, dims=('x', 'y', 'time'))
<xarray.DataArray (x: 3, y: 4, time: 2)>
array([[[0.006194, 0.380531],
[0.561032, 0.360739],
[0.515117, 0.300988],
[0.650297, 0.560798]],
[[0.703038, 0.762046],
[0.703794, 0.467468],
[0.19292 , 0.395919],
[0.344915, 0.874793]],
[[0.570962, 0.881131],
[0.758426, 0.582632],
[0.419739, 0.634596],
[0.720827, 0.12582 ]]])
Dimensions without coordinates: x, y, time
da = xr.DataArray(arr,
dims=('x', 'y', 'time'),
coords={'x': [10, 20, 30],
'y': [0.3, 0.7, 1.3, 1.5],
'time': [datetime.datetime(2016, 3, 5),
datetime.datetime(2016, 4, 7)]})
da
<xarray.DataArray (x: 3, y: 4, time: 2)>
array([[[0.006194, 0.380531],
[0.561032, 0.360739],
[0.515117, 0.300988],
[0.650297, 0.560798]],
[[0.703038, 0.762046],
[0.703794, 0.467468],
[0.19292 , 0.395919],
[0.344915, 0.874793]],
[[0.570962, 0.881131],
[0.758426, 0.582632],
[0.419739, 0.634596],
[0.720827, 0.12582 ]]])
Coordinates:
* x (x) int64 10 20 30
* y (y) float64 0.3 0.7 1.3 1.5
* time (time) datetime64[ns] 2016-03-05 2016-04-07
da.sel(time='2016-03-05')
<xarray.DataArray (x: 3, y: 4)>
array([[0.006194, 0.561032, 0.515117, 0.650297],
[0.703038, 0.703794, 0.19292 , 0.344915],
[0.570962, 0.758426, 0.419739, 0.720827]])
Coordinates:
* x (x) int64 10 20 30
* y (y) float64 0.3 0.7 1.3 1.5
time datetime64[ns] 2016-03-05
da.isel(time=1)
<xarray.DataArray (x: 3, y: 4)>
array([[0.380531, 0.360739, 0.300988, 0.560798],
[0.762046, 0.467468, 0.395919, 0.874793],
[0.881131, 0.582632, 0.634596, 0.12582 ]])
Coordinates:
* x (x) int64 10 20 30
* y (y) float64 0.3 0.7 1.3 1.5
time datetime64[ns] 2016-04-07
da.sel(x=slice(0, 20))
<xarray.DataArray (x: 2, y: 4, time: 2)>
array([[[0.006194, 0.380531],
[0.561032, 0.360739],
[0.515117, 0.300988],
[0.650297, 0.560798]],
[[0.703038, 0.762046],
[0.703794, 0.467468],
[0.19292 , 0.395919],
[0.344915, 0.874793]]])
Coordinates:
* x (x) int64 10 20
* y (y) float64 0.3 0.7 1.3 1.5
* time (time) datetime64[ns] 2016-03-05 2016-04-07
da.mean(dim='time')
<xarray.DataArray (x: 3, y: 4)>
array([[0.193362, 0.460886, 0.408053, 0.605548],
[0.732542, 0.585631, 0.294419, 0.609854],
[0.726047, 0.670529, 0.527168, 0.423323]])
Coordinates:
* x (x) int64 10 20 30
* y (y) float64 0.3 0.7 1.3 1.5
da.mean(dim=['x', 'y'])
<xarray.DataArray (time: 2)> array([0.512272, 0.527288]) Coordinates: * time (time) datetime64[ns] 2016-03-05 2016-04-07
PM25.sel(time='2014').groupby('time.month').std(dim='time')
<xarray.DataArray 'data' (month: 6, y: 1162, x: 1240)>
array([[[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]],
[[ nan, nan, ..., nan, nan],
[ 0. , nan, ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]],
...,
[[ 23.97684, 0. , ..., nan, nan],
[ 1. , 2.5 , ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]],
[[146.48776, 125.04899, ..., nan, nan],
[146.27904, 148.14934, ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]]], dtype=float32)
Coordinates:
* y (y) float64 1.429e+06 1.428e+06 1.427e+06 1.426e+06 1.424e+06 ...
* x (x) float64 -9.476e+05 -9.464e+05 -9.451e+05 -9.439e+05 ...
* month (month) int64 1 2 3 4 5 6
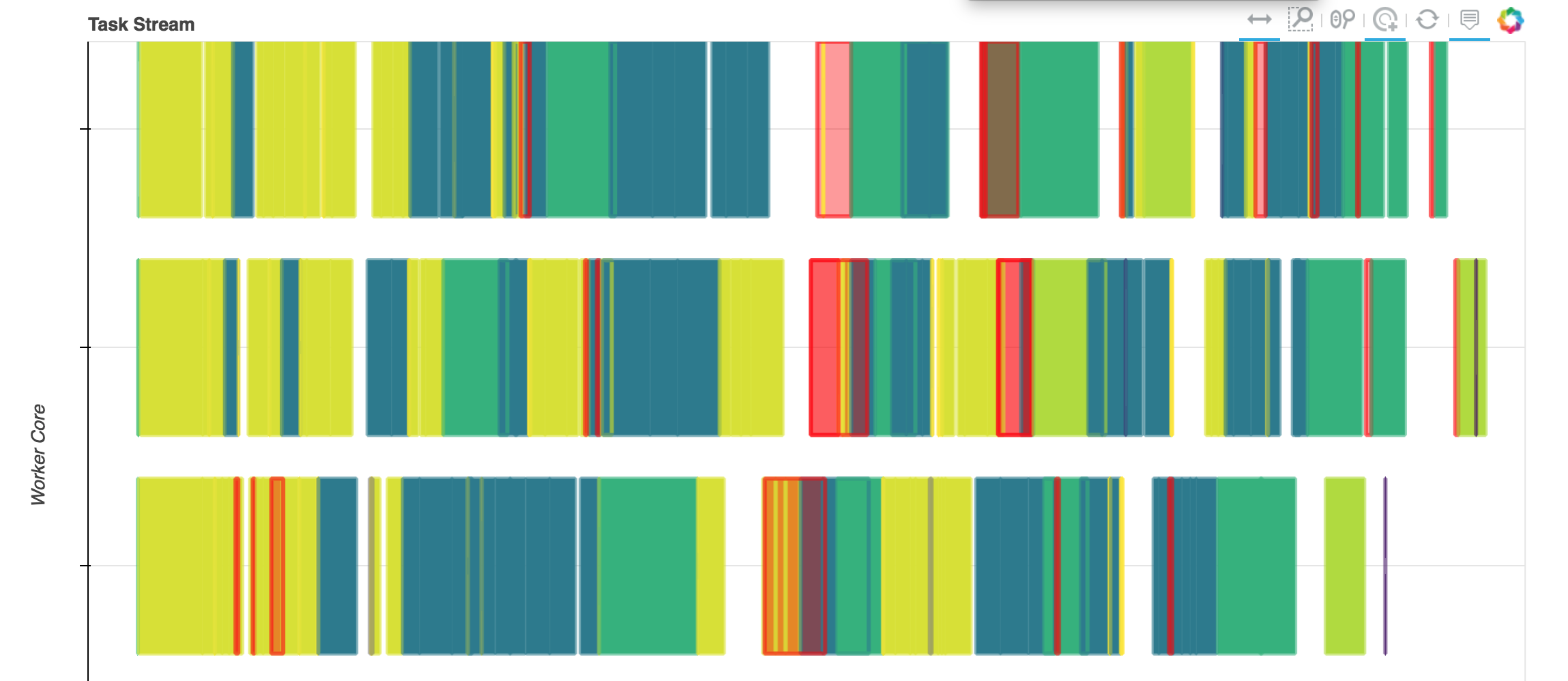
Efficient processing with dask and dask.distributed¶
dask creates a computational graph of your processing steps, and then executes it as efficiently as possible.
This includes only loading data that is actually needed and only processing things once.
data = xr.open_mfdataset(['DaskTest1.nc', 'DaskTest2.nc'], chunks={'time':10})['data']
avg = data.mean(dim='time')
seasonal = data.groupby('time.season').mean(dim='time')
dask.distributed¶
All of these different chunks, and separate processing chains can be run on separate processes or separate computers.
Getting data into xarray¶
xarray can read various formats directly:
- NetCDF
- HDF
- GRIB
xarray can now use rasterio to read loads of geographic raster formats
xr.open_rasterio('satellite_image.tif')
<xarray.DataArray (band: 3, y: 1644, x: 1435)>
[7077420 values with dtype=uint8]
Coordinates:
* band (band) int64 1 2 3
* y (y) float64 1.484e+05 1.484e+05 1.484e+05 1.484e+05 1.484e+05 ...
* x (x) float64 4.301e+05 4.301e+05 4.302e+05 4.302e+05 4.302e+05 ...
Attributes:
transform: (10.0, 0.0, 430130.2396, 0.0, -10.0, 148415.8755, 0.0, 0.0, ...
crs: +ellps=airy +k=0.999601 +lat_0=49 +lon_0=-2 +no_defs +proj=t...
res: (10.0, 10.0)
is_tiled: 0
nodatavals: (nan, nan, nan)
Example - combining multiple satellite data files into a time-series¶
Take a large number of files, one from each orbit of a satellite, and put them into one large array with a time dimension.
def file_to_da(filename):
da = xr.open_rasterio(filename)
time_str = os.path.basename(filename)[17:-17]
time_obj = datetime.datetime.strptime(time_str, '%Y%j%H%M')
da.coords['time'] = time_obj
return da.isel(band=0)
list_of_data_arrays = [file_to_da(filename) for filename in files]
combined = xr.concat(list_of_data_arrays, dim='time')
combined.shape
(10, 1162, 1240)
combined.coords
Coordinates:
band int64 1
* y (y) float64 1.429e+06 1.427e+06 1.426e+06 1.425e+06 1.424e+06 ...
* x (x) float64 -9.47e+05 -9.458e+05 -9.445e+05 -9.432e+05 ...
* time (time) datetime64[ns] 2003-04-16T14:40:00 2002-03-19T11:20:00 ...
Getting raster data out of xarray¶
from xarray_to_rasterio import xarray_to_rasterio
mean = combined.mean(dim='time', keep_attrs=True)
xarray_to_rasterio(mean, 'Mean.tif')
A few tasters...¶
Interpolation¶
PM25.interp(x=318193.5, y=176849.7).to_pandas().dropna().head()
time 2014-01-07 74.390257 2014-01-09 42.588762 2014-01-19 84.464250 2014-02-07 37.803606 2014-02-11 67.395553 dtype: float64
PM25.interp(x=318193.5, y=176849.7, method='nearest').to_pandas().dropna().head()
time 2014-01-07 72.0 2014-01-09 42.0 2014-01-19 82.0 2014-01-27 159.0 2014-02-01 297.0 dtype: float32
Resampling time series¶
PM25.resample(time='1M').mean(dim='time')
<xarray.DataArray 'data' (time: 6, y: 1162, x: 1240)>
array([[[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]],
[[ nan, nan, ..., nan, nan],
[ 20. , nan, ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]],
...,
[[ 70.333336, 46. , ..., nan, nan],
[ 49. , 53.5 , ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]],
[[333. , 326.5 , ..., nan, nan],
[324.33334 , 334.33334 , ..., nan, nan],
...,
[ nan, nan, ..., nan, nan],
[ nan, nan, ..., nan, nan]]], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2014-01-31 2014-02-28 2014-03-31 ...
* y (y) float64 1.429e+06 1.428e+06 1.427e+06 1.426e+06 1.424e+06 ...
* x (x) float64 -9.476e+05 -9.464e+05 -9.451e+05 -9.439e+05 ...
Rolling windows¶
PM25.rolling(time=5).mean()
<xarray.DataArray (time: 10, y: 1162, x: 1240)>
array([[[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan],
...,
[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan]],
[[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan],
...,
[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan]],
...,
[[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan],
...,
[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan]],
[[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan],
...,
[nan, nan, ..., nan, nan],
[nan, nan, ..., nan, nan]]], dtype=float32)
Coordinates:
band int64 1
* y (y) float64 1.429e+06 1.427e+06 1.426e+06 1.425e+06 1.424e+06 ...
* x (x) float64 -9.47e+05 -9.458e+05 -9.445e+05 -9.432e+05 ...
* time (time) datetime64[ns] 2003-04-16T14:40:00 2002-03-19T11:20:00 ...
OPeNDAP¶
Accessing data over the internet - but only downloading the bits you use
dataset = xr.open_dataset('http://opendap.knmi.nl/knmi/thredds/dodsC/e-obs_0.25regular/tg_0.25deg_reg_v17.0.nc')
dataset['tg']
<xarray.DataArray 'tg' (time: 24837, latitude: 201, longitude: 464)>
[2316397968 values with dtype=float32]
Coordinates:
* longitude (longitude) float32 -40.375 -40.125 -39.875 -39.625 -39.375 ...
* latitude (latitude) float32 25.375 25.625 25.875 26.125 26.375 26.625 ...
* time (time) datetime64[ns] 1950-01-01 1950-01-02 1950-01-03 ...
Attributes:
long_name: mean temperature
units: Celsius
standard_name: air_temperature
temperature = dataset['tg']
oneday = temperature.sel(time='2009-07-01')
oneday.plot(robust=True)
<matplotlib.collections.QuadMesh at 0x324b65080>
xarray extensions¶
Simulation models in xarray (
xarray-simlab)WRF Weather Forecasting Model functions (
wrf-python)Empirical Orthogonal Functions (
eofs)Many more at http://xarray.pydata.org/en/stable/faq.html#what-other-projects-leverage-xarray
from eofs.xarray import Eof
monthly = PM25.resample(time='M').mean('time')
solver = Eof(monthly)
results = solver.eofs()
results.plot(col='mode', col_wrap=3, robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x324a77550>
Resources¶
Slides: http://bit.do/xarray_pyconuk
Code: https://github.com/robintw/XArray_PyConUK2018
XArray docs: http://xarray.pydata.org/
robin@rtwilson.com @sciremotesense @robintw on Slack